23andMe lets you download a spreadsheet with details of your DNA relatives, including the chromosomes and positions where you share segments of DNA.
This is very helpful for several tasks, including triangulating your relatives to research common ancestors. If you haven’t tried triangulation, it’s probably a lot easier than you think.
I use the spreadsheet to pick and choose groups of DNA relatives to start the process. This article is a step-by-step walkthrough.
Download Your DNA Relatives Spreadsheet
23andMe provides a file with all your DNA relatives. The information includes
- which chromosome(s) have the shared DNA
- the size of each section of shared DNA
- the start and end positions of the shared segment on the chromosome
To download the file, open the DNA Relatives list and scroll to the bottom of the page.
You’ll see a button for requesting the file.

23andMe will send you an email link when your file is ready.
Their instructions say that there could be a wait of 24 hours. It took five minutes for me from my most recent request.
Convert The DNA Relatives File To A Spreadsheet
23andMe gives you a zip file that contains what’s known as a CSV file. You should open this file with your preferred spreadsheet tool.
I like to use Microsoft Excel. You may want to open the file with Google Sheets. The process I’ll describe now is very similar, no matter what tool you use.
Using Excel, I save the file in .xlsx format to get access to the spreadsheet features.
If you upload the file into Google Sheets, the online software will convert the data into spreadsheet format.
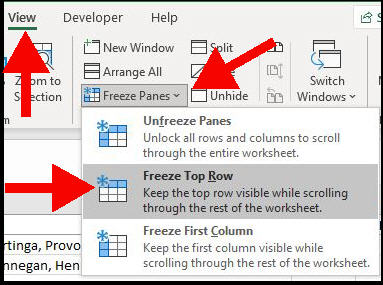
Freeze The Top Row
The first thing I do is to freeze the header row.
This is because I will be changing the sort order of the data, and doing a bit of scrolling up and down.
Sort The Spreadsheet By Chromosome And Position
The spreadsheet is sorted in the same way as your DNA Relative List. Your relatives are listed in descending order of the closeness of relationship with you.
My top relative has two rows in the spreadsheet. This is because we share DNA on two different segments on the 7th and 16th chromosome pair.
One of my other relatives has five rows. Two shared segments are on the 2nd chromosome pair, and the rest are on three different chromosomes.
But this isn’t the sort order that I want for how I will use the spreadsheet. So, my next step is to change how it’s sorted.
You probably know how to order a spreadsheet by a single column. In this case, we want to use three columns:
- Chromosome #
- Start position
- End position
In Excel, you achieve this by choosing the “Custom Sort” option from the Sort & Filter menu. This lets you select the three columns in the order I list them above.
Hide Unwanted Columns
This step is optional. But I prefer an uncluttered display where I can see what I need without horizontal scrolling across the screen.
These are the columns that I hide:
- Surname
- # SNPs
- Full IBD
- Sex
- Set relationship
- Predicted relationship
- Relative range
- Maternal
- Paternal
- Maternal haplogroup
- Paternal haplogroup
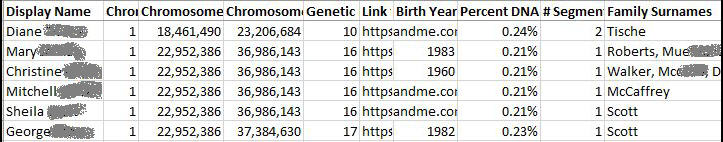
This means that I can see all the information I want with one glance, including the family surnames.
Extra Formatting
This step is also optional, but I think it makes the spreadsheet much easier to deal with.
The chromosome start and end positions are lengthy numbers. I find them easier to read when they are in the “1000,000,000” format i.e. commas separate the thousands.
So, I change the formatting of these two columns to display as a Number with the 1000 separator.
The Genetic Distance column shows the number of centimorgans with five decimal places. Again, this is awkward to read. I format the column to display as a round number.
Here’s an excerpt from my spreadsheet. My point is that I can see the name, chromosome details, relationship (cMs and percentage), and family surnames at a glance.
Find The Segment for Your Target Relative In The Spreadsheet
If you don’t already have a target to investigate, you may as well start with your closest relative with whom you have not yet identified a connection.
For me, that was my highest match on 23andMe. This relative, named Jennifer, is described as a 3rd cousin.
My first step with unknown relatives is to open their profiles and look at our Relatives In Common.
In Jennifer’s case, we have plenty of fourth cousins in common and I don’t know where any of them fit in my family tree.
But if I can figure out my connection with Jennifer, a lot of other relatives could fall into place.
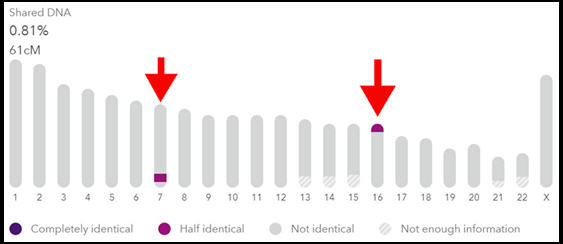
When I expand the DNA details for Jennifer, the chromosome browser on this page tells me we share segments on two different chromosomes.
I’ve got a separate article that runs through using the one-to-one chromosome browser on 23andMe
But I don’t know which segment is longer than the other yet. One drawback of this version of the browser on 23andMe is that it doesn’t show the different segment sizes.
So, my next port of call is to the spreadsheet. I just need to remember that Jennifer will appear several times (one row per shared segment).
I use the “find” feature in the spreadsheet to search for Jennifer. She appears twice in the spreadsheet.
Now, I choose a segment (usually the largest) and take a look at the neighboring rows.
Identify Other Relatives Who Share This Segment
The reason why we ordered the spreadsheet this way is that it makes it easy to identify a group of relatives who share the same overlapping segments.
In the excerpt below, notice how Jennifer’s segment has the exact same end position (27,377,120) as Derek, the guy below. But they have different starting positions.

While Ryan and Joan (the first two rows) don’t match either the starting or end positions of Jennifer. Although they themselves have the same starting position as each other.
Look for overlap, not alignment
Don’t think that your group of DNA relatives need to be aligned on the same starting and end position. That will rarely happen.
But the overlapping sections need to be of significant length. Otherwise, you could be dealing with sections that are identical by chance.
Eliminating Close Relatives
I’ve obscured some details of these matches, but I can tell you that Jennifer and Derek have listed the same four names in their “Family Surnames” section.
That was a bit of a clue! Having the same endpoint made me also wonder about how closely these two people were related to each other.
However, when I looked at the Relatives In Common list for Jennifer, there was no sign of Derek on the first page of our shared relatives. I had to go to the fifth page to find Derek. Sure enough, he is labeled as her brother.
One of the drawbacks of the Relatives In Common list is that you can’t change the sort order. It is ordered by how close the relationship is with you, not the DNA match.
My shared percentage of DNA with Jennifer is 0.81% while Derek is a fraction of that at 0.24%
It’s down to the random nature of inheritance that I share over three times the amount of DNA with Jennifer as with her sibling!
However, I’ve now identified that two of the people in my group are siblings. I’m going to remove Derek, who shares less DNA with me. But why?
Because for the purpose of triangulation, having Derek doesn’t give extra value over Jennifer. This is because they have an identical pedigree (parents, grandparents, etc).
So, it’s worth eliminating one sibling and bringing in another row from the spreadsheet that overlaps with this area.
Close relatives are useful in other ways
That doesn’t mean you should now ignore the interesting fact that these two people are close relatives.
One may be much more open to communication than the other. One may have a tiny family tree on another genealogy website, while the sibling has a much bigger version.
I add notes to 23andMe and to the spreadsheet to remind myself of this.
Repeating the process of elimination
When I looked for another row to add to the Chromosome Browser, I skipped two who had the same surname as Ryan.
But before I skipped them, I looked at my Relatives In Common with Ryan. I had to go as far as page four to find that these two were Ryan’s sister and mother.
More useful information from close relatives
I also noticed that Ryan was 1st cousin to Kathleen and also to another row in the spreadsheet in the Relatives In Common list.
So, I worked through my Relatives in Common with Kathleen to find that Kathleen was a sister to this lady.
Kathleen hadn’t added any family surnames, but her sister had.
I didn’t swap Kathleen out of the browser for her sister. But I added the sister’s family surname details into Kathleen’s row in the spreadsheet (with an explanatory note to remind myself why).
Swapping in more relatives into the group
My goal is to get five people in the group who are distant cousins to each other, and overlap on the same chromosome.
The reason for the magic number five is that I will be using the Advanced Chromosome Browser in a later step. This browser allows a maximum of five DNA relatives to compare to your own kit.
Next Steps – Triangulation
This article was about forming a group of your DNA relatives that have overlapping DNA on the same chromosome.
Identifying common ancestors
The end goal is to ensure that every member of the group inherits this piece of DNA from a common ancestor.
This lets you switch to standard genealogy techniques to look for intersections of surnames and locations in their family trees.
You may be able to gather this information through communicating with your DNA relatives. Alternatively, you may be able to find their public trees on other platforms. And you may have to roll up your sleeves and build back their family trees for them!
Triangulation
The challenge is that at this point of your preparation, you cannot be sure that everybody in the group shares the same piece of DNA with each other.
All you know now is that they share the section with you.
Triangulation is the process that takes you to the end goal. On 23andMe, you can use a combination of the Relatives In Common display and the advanced chromosome browser.
I’ve got a separate article that walks through the process of this technique. Consider it as part two to the article you’re reading now.
So, your next step is to work through the steps in our article on triangulation with the 23andMe chromosome browsers.
Thank you for this article. It is very helpful and I will try to use it to help with my matches. I use LibreOffice, but think my issues with this spreadsheet might be similar if using Excel to view it. I find the original 23andMe relatives spreadsheet formatting to be frustrating for several reasons. But maybe I’m being too “nit-picky?”
One example: Some matches will show a lot of information, such as multiple surnames and locations. If a match shows these multiples, each surname or location is put in a separate cell, and that throws the column titles off for the ensuing content for that line/match. Subsequent surnames after the first will be in the columns for Family Locations, Maternal Grandmother Birth Country, and so on. Experimenting with one such match, I was able to merge all the surname cells into the first one, but then had problems trying to wrap that text. Then the other columns remained, so needed deleting. This would be too time-consuming to do for the whole list. Is there a better way to do it (you can explain in Excel terms, and I will try to convert to LibreOffice)? And why do the 23andMe spreadsheets do this assigning of each surname to a separate cell?
Another irritant is the column headings, which sometimes have long names but the corresponding cells will have very short text/numerals for information. To be able to condense the width of the spreadsheet, I like to “tighten up” such columns by selecting the column heading cell and enabling wrapping, but then it takes some fidgeting to get the headings centered. I know I should just live with this, or hide columns to help condense.
If you have any advice for the above, I would appreciate it.
BONUS: Many people manage several DNA profile kits (self, parent, grandparent, 1st cousin, great-aunt/uncle, 2nd cousin). By downloading and including these other matches (add an extra column to label the data source/person), one can make a more robust overlap/triangulation tool. One could go further and flag specific segments of DNA and attribute this to a specific ancestor branch (home-made-Lazarus-tool).
Also:
1) there are formulas in excel for overlap.
2) create a secondary table for the TOTAL shared cM per match to indicate there are more/other locations of shared DNA. vlookup this back into the base data table.
3) advanced: Use graphical overlay of data; Use formulas to match your logic/process for identifying target matches…. stop scrolling.
Thanks for taking the time to give us these extra ideas.