GEDmatch accepts DNA tests from all the major consumer DNA testing companies. Their free comparison tools allow you to compare your uploaded DNA with a collection of archaic DNA matches.
These ancient samples come from fossilized remains found across the world. Several of the samples are Neanderthal DNA.
The Archaic DNA Matches Tool shows you which segments of DNA you share with these ancient samples.
The One-To-One Comparison Tool lets you pick an archaic DNA match and see a more detailed breakdown.
So, how do you go about using these tools? And when you find shared DNA, what does this actually mean?
This article provides an explanation of ancient matches on GEDmatch, and a step-by-step guide on using the comparison tools.
What Are The Neanderthal and Other Archaic DNA Matches On GEDmatch?
These are the best known Neanderthal and related samples:
- Altai Neanderthal
- Mezmaiskaya Neanderthal
- Denisova
Altai Neanderthal
The Altai Neanderthal kit represents DNA from a woman who died at least 50 thousand years ago.
Some of her fossilized remains were found in Siberia in the Altai Mountains.
Mezmaiskaya Neanderthal
Other kits are for Neanderthals found in archaeological sites across Russia.
Three Neanderthals were found in the Mezmaiskaya Cave in the Caucasus Mountains.
One was an infant, whose skeleton was almost intact. The DNA kit labeled as “Mezmaiskaya N.” is from that infant.
Denisova
The Denisova Cave in the Altai Mountains also yielded DNA samples from a female who died at least 30 thousand years ago.
Genetic scientists concluded that her DNA was related to Neanderthal DNA, but represents at least a differing sub-species.
This is now called Denisovan DNA.
Other archaic samples
The archaic collection has many more kits other than Neanderthal.
People with Native American heritage may find the Clovis sample very interesting. This is from an infant boy found in Montana, whose remains date back over twelve thousand years.
And there are many samples from across Europe.
The trick is to see which kits you share the largest segments and do a bit of background research on those samples.
How Did GEDmatch Get Neanderthal and Other Archaic DNA Samples?
Are you wondering if GEDmatch users donned Panama hats and ventured into Siberian caves in search of Neanderthal bones? The answer is no.
The research scientists investigating the DNA samples put their genome files into the public domain.
A genealogy enthusiast translated those files into the format utilized by autosomal DNA testing companies like Ancestry and 23andMe. He kindly uploaded his collection to GEDmatch.
Felix Immanuel is the name of the volunteer. I found it a little difficult to research this article until I realized that Felix had changed his last name for religious reasons.
If you’re also running online searches for this topic, you will find references to his work under Chandrakumar as well as Immanuel.
Using Low Thresholds of Shared DNA With Archaic Matches
If you’ve already been using your DNA matches for family research, then you probably know that lower amounts of shared DNA may be very misleading.
Due to the random nature of inheritance, two DNA samples may have common pieces of autosomal DNA by chance.
What is autosomal DNA? If you’ve tested with Ancestry, 23andMe, MyHeritage, or LivingDNA – your DNA results are autosomal DNA.
Only FamilyTreeDNA offers the alternative Y-DNA and mitochondrial DNA tests. For this article, we’ll ignore these other two types.
IBC – Identical By Chance
Identical by Chance means that the shared DNA is not due to a common ancestor i.e. you are not related to your DNA match. Consider these to be false positives.
Of course, it would be a great waste of your time if you spent hours reviewing the family tree of a “false positive” DNA match.
The consumer DNA companies set minimum thresholds below which they will not show you DNA matches.
Different companies have different thresholds. Ancestry’s current threshold is 8 cM.
But you are not going to share enough DNA with an archaic sample to meet that kind of threshold. You have to drop lower than 8 cM. This is what GEDmatch allows you to do.
But the further you go down – the more likely it is that very small pieces of shared autosomal DNA are by chance.
However, there is still the possibility that some are not due to chance. What are the other reasons? There are two.
IBD – Identical by Descent
Identical by Descent means that the shared DNA has been inherited from a common ancestor. This is what we are usually researching.
Higher amounts of shared DNA point to an ancestor within more recent times. So, we are more likely to be able to identify the relationship with documentary evidence.
There is one piece of confusion that I see pop up in genealogy forums when discussing archaic DNA matches. Some of the samples are from children.
So the questioner asks – how can anyone possibly descend from an infant?
Well, we couldn’t. The possibility is that some of us share a common ancestor with the child, and thus received the same small piece of DNA.
But how do we know if that’s the case? The fact is that with current technology: we don’t know. That may change with future genetic analysis.
Besides, there is another possibility for shared DNA.
IBP – Identical By Population
Some pieces of DNA are prevalent within a grouping of people through many generations.
Yes, there will be a long-ago common ancestor, but too far back to identify. But these small pieces point to ancestral regions.
IBP matching is unlikely to be useful when researching your family tree. But that’s not the purpose of looking at your archaic DNA matches.
If you share tiny pieces of DNA with a Neanderthal woman found in the Altai Mountains – it could be chance, or it could be in part due to migration patterns.
And it’s these non-chance possibilities that are fascinating.
Fascinating and fun. One genealogy hobbyist asked in a forum – what can I tell from these small shared DNA segments?
The best answer, in my opinion, was: “you can tell that you’re human.”
So, don’t overreach for conclusions when looking at these archaic matches. Just marvel at the fact that we can actually run these comparisons in the present day.
Video Walkthrough: Research Archaic DNA Matches
This video is a walkthrough of the steps covered in the rest of this article.
How To Use the Archaic DNA Matching Tool
The next sections are a tutorial on using GEDmatch to compare your DNA to the archaic samples.
Step 1 – Transfer Your DNA to GEDmatch
If you’ve tested with Ancestry, I have step-by-step tutorials on how to get your DNA results onto GEDmatch. Take your pick from articles or video format.
| ARTICLE | YOUTUBE VIDEO |
| How To Download Your Ancestry DNA | Download Your Ancestry DNA |
| How To Upload Your DNA To GEDmatch | Upload Your DNA to GEDmatch |
Step 2 – Launch the “Archaic DNA Matches” Application
You’ll find the link near the bottom of the DNA Applications menu.
Plug in your kit number (this link will show you where to find it).
Then before you go any further, I suggest that you change the default setting for the segment setting. The default upper limit is set to 0.5 cM.
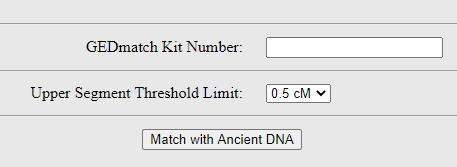
That is incredibly low. Instead, it’s interesting to work your way down from a higher setting where you have no matches. And see at which point do the matching DNA pieces pop up.
I suggest starting at 5 cM, and work down from there.
Step 3 – (First Time) Raise The Upper Segment Threshold Limit
At 5 cM, I get the message “No matching segments. Try a lower threshold”.
Hit the back button, and drop down to 4. If you get the same blank display, drop down another notch.
Step 4 – Find Your Largest Archaic Match(es)
For me, my hits kick in at 3 cm. I have shared segments with a three thousand and a seven thousand-year-old specimen. But way older is the 45 thousand male thigh bone found near the Ust-Ishim village of Omsk, Siberia.
When you get a match, you’ll probably want more information about the sample.
Step 5 – Investigate Your Largest Archaic DNA Matches
Because GEDmatch is so minimalist in its approach to helpful documentation, I’ll break down the display of my Ust-Ishim match.
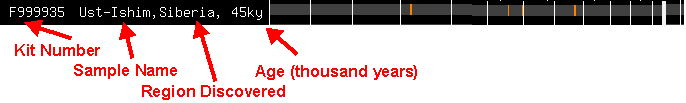
It starts with a GEDmatch Kit Number. F999935, in this case. Why do all the Kit numbers start with F? I assume that’s for their uploader, Felix.
Then comes the sample name or identifier, provided by the scientific community.
When you throw the sample name into your search engine, you’ll get scientific articles, wiki pages, and blogs. Yes, even “RISE174” will give you plenty of rabbit holes to fall down.
The identifier is followed by the area in which the sample was found. And finally, the estimated age of the sample in millennia (i.e. thousand years).
Supporting Documentation
Unfortunatelty, the researcher’s website is no longer online. That means that the supporting documentation is also gone.
The GEDmatch results page gives you a link to a document in the WayBack Machine (the Internet Archive of lost pages).
I’ve republished the list of archaic samples on GEDmatch on this website. I’ve also added an extra link to more info about each sample.
This full list is useful, because you don’t seem to see all the samples in the GEDmatch display. At least, I don’t.
If you watch our companion video to this article, you’ll see my confusion! I think the display excludes samples that you don’t even match at 0.5 cm. I may be wrong – I couldn’t find any other explanation.
Step 6 – Keep Going Down To Lower cM Thresholds
Are you a little disappointed not to see anything vaguely Neanderthal yet? Just keep working down the threshold and see an expanding list of matching DNA segments.
If you have recent European lineage, the samples found across Europe are going to start lighting up the board.
Look for the contrasts between samples with many matches, and samples with few bars. Then think about the region and time period.
Is it all a little confusing? Yes.
But fascinating? Yes.
Do I have any particular insights for you? No…other than we’re all human.
Step 6 – Are You Particularly Interested In Neanderthal DNA? Look For These Samples
For me, a setting of 1 cM gives me matching segments with “Altai N.”, the Neanderthal woman found in the Altai Mountains.
She is one of the first named samples, so she’ll be at the top of your display when you see her. Right beneath is the Denisovan sample.
But which of the other samples are Neanderthal? Some have a sample name that is clearly Neanderthal, but not all.
The list below is an extract from the supporting document of all samples with Neanderthal as part of the name.
- Altai Neanderthal
- Mezmaiskaya Neanderthal
- Feld1 Neanderthal
- Sid1253 Neanderthal
- Vi33.16 Neanderthal
- Vi33.25 Neanderthal
- Vi33.26 Neanderthal
I’m not sure if that represents all the Neanderthal samples.
Using The GEDMatch One-To-One DNA Comparison With Archaic DNA Matches
Remember that all the DNA matches listed in the Archaic display are simply GEDmatch DNA kits. Just like the kits belonging to you and every other GEDmatch user.
That means you can use some of the other tools on GEDmatch with these archaic kits. For example, you can use the One-To-One Autosomal DNA Comparison tool to compare any single kit to your own.
Personally, I have to drop to 3 cM to get my first few archaic DNA matches. That happens to be the minimum threshold for the One-To-One comparison tool. So, I’m limited to the number of archaic samples I can compare with.
But here’s a step-by-step guide for the One-To-One Comparison.
Step 1: Launch the One-To-One Comparison Tool
The One-To-One tool is the third item in the DNA Applications menu on the home page.
Step 2: Choose an Ancient Kit Number
This is where you enter both your kit and the kit number for one of the ancient samples.
You have to type in the archaic kit numbers from the display in the Archaic Comparison. As the display is an image, unfortunately you can’t just copy the text. For repeated use, type the samples of interest into a little text file for re-use.
For convenience, here are two of the Neanderthal samples as well as the Denisova sample.
- F999902 Altai Neanderthal
- F999909 Mezmaiskaya Neanderthal
- F999903 Denisova
Step 3: Set The Minimum CM Threshold
The minimum segment cM size is set to 7 by default. So, you will have to lower this threshold to match with the archaic samples. You can’t lower it below 3 cM, but hopefully this is enough for your matches of interest.

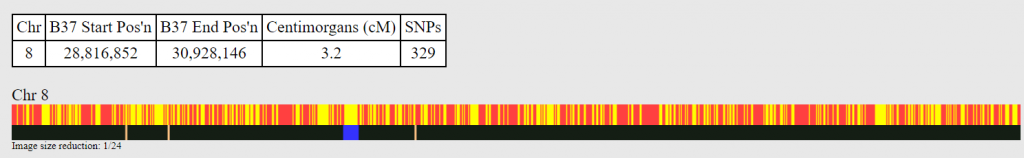
Here is my 8th chromosome match with the 45 thousand-year-old Ust-Ishim Siberian sample.
Ancient Irish DNA Matches
There is a separate set of Irish archaic samples that were uploaded by another researcher. So, they’re not found with the Archaic Matches we’ve shown in this article.
I’ve got a separate article on researching ancient Irish DNA matches on GEDmatch.
My Conclusion About Archaic DNA Matches: Fascinating and Fun
If you followed along with this walkthrough, I hope you found it to be as interesting as I did.
The GEDmatch uploads were provided in 2014. If you choose to read up on the Altai and Denisovan discoveries, you’ll find that the scientific analysis and insights are fast-moving and changing even since then.
I’m very conscious of the dangers of working with such centimorgan thresholds, so I’ll only draw this conclusion: examining these archaic matches has been both fascinating and fun.

i have one hit at 7cm F999941 rise 98 sweden 3.7ky chr 16 looks like
mother gets a hit at 6cm 999937 NE1 Hungary 7.2ky chr 15
Thanks for your video! It was helpful–
Quite randomly, I had chosen a random archaic cousin to look up an it was one of the Croatian Neanderthals from the Vindija Cave: Vi33.26!
So yes, it does show up for me…..
Thank you for this very clear tutorial
I saw a huge list of ancient samples that he left out, is there anyway to see them?
Are you referring to his original list on his website? I noticed that it is no longer live. GEDmatch give a link to the WayBack Machine to find the page.
I’ve republished the list of GEDmatch archaic samples here, with some extra info.